Highly enhanced mutagenesis method for generating large high-diversity gene libraries
Background
Gene libraries are an invaluable tool for recombinant protein development and engineering. The size and quality of the libraries are, however, typically limited by bottlenecks in the mutagenesis and transformation processes.
Description
Large gene libraries with high mutant frequency are generated using a highly controllable, enzyme-dependent selection of mutated strands in primer extension mutagenesis using a strand-displacing DNA polymerase.
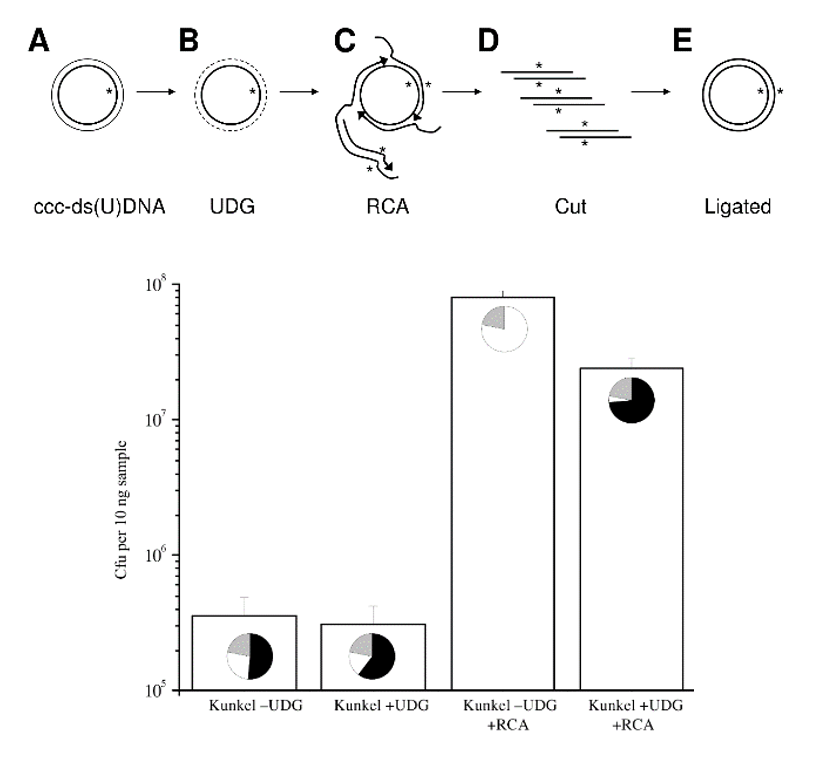
Starting with few hundred nanograms of heteroduplex DNA, billion-member gene libraries can be achieved by using uracil-DNA glycosylase treatment and selective rolling circle amplification (sRCA) on uracil-containing DNA templates, allowing:
- Selective amplification of mutated strands only
- Mutagenesis efficiency close to 100%
- Nearly 300-fold increased number of transformants per nanogram of source DNA
- Transformed cells only contain mutated DNA
- Suitable for mutating both single-stranded and double-stranded DNA
- Tens of large libraries can be generated in parallel
- All DNA handling can performed in microtiter wells.
Application Areas
Development and engineering of recombinant proteins by oligonucleotide targeted random mutagenesis with significant improvements. Enhancing throughput of affinity maturation.
The IP is available for licensing from the TTO of the University of Turku (UTU)
ID: UTU150201
Title: MUTAGENESIS METHOD
PCT Publication: WO2011077004 (A1)
Granted patents: EP2516643 (B1), CN102695795 (B), US9453216 (B2)
Status: Technology Readiness Level (TRL) 7
Well optimized and validated method. Notable interest from the scientific community with independently demonstrated libraries using the technique. Significant improvements in affinity maturation process reported.